Steering Cells
The paper: Dang, et al. (2013) Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503: 281-284. doi:10.1038/nature12611
Subject areas: Cell Biology
Vocabulary:
cytoskeleton – cells are not just completely fluid structures surrounded by a flexible membrane. Cells must have at least some rigidity in order to generate force so that they can either move from one place to another, or at least to move materials within the cell from one place to another. The cytoskeleton refers to the structural proteins that accomplish that task: in eukaryotes, the main kinds of cytoskeletal proteins are the actin microfilaments mentioned here, the slightly larger intermediate filaments, and the microtubules.
—–
This article is a summary of a recent primary research paper intended for high school teachers to add to their general knowledge of current biology, or to supplement their lessons by showing students the kinds of projects that current biological research addresses.
—–
In some ways, this is exactly the kind of article that at first glance would cause casual readers to skip over it. It has a ridiculous number of authors (34!), and a quick glance at the opening few sentences brings up this sixteen-word phrase “Wiskott–Aldrich syndrome protein (WASP)-family verprolin-homologous protein (WAVE, also known as SCAR)” simply to name a protein. All that aside, this article addresses a very important cellular mechanism: locomotion.
When you think about most of the cells of your body, you think of them as stationary, anchored in place relative to their neighboring cells, other than a few exceptions. That is true, but cell motility plays some very important physiological roles, such as in development as cells need to extend or migrate from their “birthplace” to their eventual adult position, or in wound healing when a variety of cell types move to the wound to begin the repair work if there is an injury, or even pathologically, as a crucial step in the spread of cancer beyond an initial tumor. Of course, these kinds of cell movement can also apply to cells from other organisms, even simple single-celled amoebae.
For cells moving across a solid surface, one means of locomotion is to extend the leading edge of the cell membrane in the direction desired, grip the surface, and pull the rest of the cell body towards it. It’s not much different than taking a step, having your foot or shoe grip the floor, and the muscles of your body coordinate to move it forward. However, in the case of the cell, the membrane itself has no capability for movement, and it is essentially amorphous. So, what needs to happen is a coordinated rearrangement of the cytoskeleton that accomplishes the extension of that leading edge of membrane.
The extending leading portions of a cell membrane are known as lamellipodia. Although the basic idea of cytoskeletal rearrangement to carry out lamellipodial extension is not new, there are still many holes in our knowledge of both control and motor mechanisms. This paper addresses one of those outstanding issues.
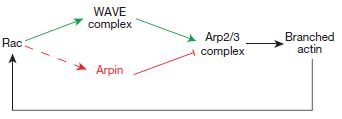
It was already known that one of the mechanisms is the generation of new actin filament networks that push the membrane outward as they are formed. The Arp2/3 complex is also known to be involved in this by nucleating or initiating the formation of filaments from individual actin molecules (which are roughly spherical). Arp2/3 is activated by WAVE, which would have been activated by Rac, which is one of the primary initiators of multiple aspects of lamellipodial formation.
As it turns out, a new protein, which the investigators have named Arpin, has been described that interacts with Rac at the tips of the lamellipodia. Interestingly, Rac activation of arpin seems to be at odds with Rac activation of WAVE.
What they knew
Arp2/3 is activated by different WASP-family proteins in different situations. At the lamellipodia tips, WAVE binds to Arp2/3 through an acidic (i.e. negatively charged) region at one end of the protein. When that acidic region of WAVE binds Arp2/3, it causes the Arp2/3 to change its conformation, or shape, which makes turns it on, allowing it to begin nucleating new actin filaments. These newly elongating filaments help to push the leading edge of the lamellipodia outward.
In many biological systems, there is a push for every pull: if there is a mechanism to induce growth, there is usually a mechanism that inhibits growth, and the balance of the two lead to proper function. In other examples of Arp2/3 there are inhibitory proteins that oppose the action of the WASPs. Since they also have similar acidic regions, they likely compete for Arp2/3 binding but do not cause the correct conformational change to activate it. Known inhibitors such as PICK1 and Gadkin have not been described in the lamellipodia.
What they did
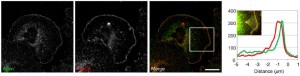
Guessing that if there was an inhibitory protein opposing WAVE in the lamellipodial tips, it would probably also have the characteristic acidic region, Dang et al did a database search for similar protein sequences. In fact, they found an uncharacterized protein, labeled C15orf38, along with a number of WASP-like nucleation factors. This seemed promising, so they decided to begin characterizing the protein. Looking at where in the cell this protein, which they called arpin, was located, they stained embryonic mouse fibroblasts for arpin, as well as WAVE, Arp2/3, and cortactin (another lamellipodial protein). From left to right, the figure below (click to expand) shows staining of arpin and the Arp2/3 complex separately, then merged.
In the merged photomicrograph, the arpin staining is green, the Arp2/3 staining is red, and areas where they are located together show up as yellow. Looking at either the photo or the graph next to it showing staining intensity at different distances from the very edge of the cell (0 on the x-axis), there is clearly some co-localization of the two proteins at the lamellipodial tips/edges of the cell. In comparison, co-localization of arpin and WAVE is even stronger and again on the edge of the extending membranes.
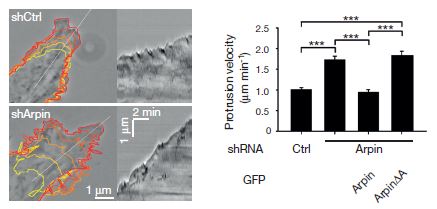
What does arpin do? Is it a nucleating factor like WAVE, or is it an inhibitory factor? They tested that by introducing inhibitory RNA molecules specific for arpin. The figure shows that inhibiting the synthesis of arpin causes the lamellipodia to protrude faster. The graph shows further that if more arpin is injected into the cells that have been depleted, the speed of the lamellipodia drops back to normal.
The next step was to examine the effects of Rac on the interaction between Arp2/3 and arpin. In the figure at right, the important 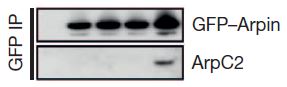 part is the bottom row, which shows that Arp2/3 is only co-precipitated with arpin when activated Rac is added. Co-precipitation is an experiment in which an antibody that specifically binds a protein of interest (in this case arpin) is stuck to a support like a gel microbead. If you run a solution extracted from a cell through a column containing many of these beads, the arpin will get stuck to the beads as most other proteins continue to run through. It is called a co-precipitation experiment because you can alter the conditions so that other proteins that are tightly stuck to arpin will remain stuck to it through this process. Then, the proteins that have been stuck to the antibody-beads can be knocked off the beads (often with high-concentration salt solution) and analyzed. Going left to right, the empty lane is a control that should not bind any arpin or Arp2/3 protein, the next lanes are no Rac, a dominant negative Rac, normal but inactive Rac, and Rac that is constantly active.
part is the bottom row, which shows that Arp2/3 is only co-precipitated with arpin when activated Rac is added. Co-precipitation is an experiment in which an antibody that specifically binds a protein of interest (in this case arpin) is stuck to a support like a gel microbead. If you run a solution extracted from a cell through a column containing many of these beads, the arpin will get stuck to the beads as most other proteins continue to run through. It is called a co-precipitation experiment because you can alter the conditions so that other proteins that are tightly stuck to arpin will remain stuck to it through this process. Then, the proteins that have been stuck to the antibody-beads can be knocked off the beads (often with high-concentration salt solution) and analyzed. Going left to right, the empty lane is a control that should not bind any arpin or Arp2/3 protein, the next lanes are no Rac, a dominant negative Rac, normal but inactive Rac, and Rac that is constantly active.
What does it mean?
So this is a bit counter-intuitive. Rac on the one hand activates WAVE, which turns on Arp2/3, which nucleates new actin filament polymerization to help push the leading edge of the lamellipodia, but on the other hand also activates arpin, which inhibits Arp2/3 thus slowing or preventing extension of the lamellipodia. What does it all add up to? It would appear that even as Rac initiates lamellipodial extensions, it has one foot on the brakes (arpin) to keep the exploration cautious and relatively slow.
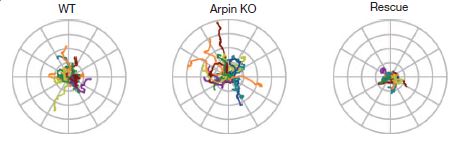
In the image here, you can see traces of the movement of the amoeba Dictyostelium discoideum as it moves around. On the left are the normal wild-type amoebae paths, in the middle are the paths from arpin-knockout amoebae, and on the right are traces of the knockout amoebae that have had arpin injected back into them. The arpin knockouts travel farther/faster and also a bit straighter for longer periods of time than the wild-type or arpin-rescued.
By modulating either the availability or activation of arpin, the cell appears to steer the formation of lamellipodia – turning more and exploring close to its position when arpin is active, but making faster runs away when arpin is not.

No comments
Be the first one to leave a comment.